Rapid Detection of Multidrug-resistant Microorganisms by Surface-enhanced Raman Spectroscopy
Bacterial infection is a frequent complication among trauma and surgical patients, both civilian and military. The rate of infectious complications in the U. S. military is approximately 35 percent for combat casualties. The lack of a rapid diagnostic platform for the detection of bacterial and fungal infections in combat wounds result in mortality and extended stays in Intensive Care Units. The methods currently available are time consuming, lack sensitivity, require specialized equipment, and highly trained personnel. There is a need for a field deployable (portable) rapid point-of-care diagnostic platform to detect and diagnose infections in a short period of time.
A technique known as surface enhanced Raman scattering (SERS) spectroscopy, has the ability to generate unique spectral biomolecular “fingerprints” of microbes such as bacteria and viruses.
This study evaluates the feasibility of utilizing a hand-held SERS based system for the detection and generation of "molecular fingerprints" of military relevant microorganisms often associated with wound infections.
A field deployable SERS diagnostic platform that has the ability to identify microorganisms within minutes, will be beneficial for the rapid identification and treatment of bacteria in combat-injured military personnel, and actually have an impact on improving trauma-related outcomes.
Navy researchers are determining if SERS allows for the generation of unique Raman spectra for the rapid identification of biological agents of military importance, leading to novel rapid diagnostic assays to replace the standard time consuming procedures currently in place.
This project is using a DXR tabletop SERS microsope, a hand held Raman spectrometer, and multi-well substrates. The efficacy of different lysis buffers to purify bacteria from complex matrices including saliva, blood and sera are being evaluated.
The spectra of each bacterial species represent shifts in the frequency of monochromatic (fixed frequency) light caused by the biological structures of the bacterial cell wall. Hydrophilic bacterial species of Staphylococcus aureus and Acinetobacter baumannii were easily detected and identified from serum samples without significant changes occurring to their spectra due to sample processing. Shifts in relative peak intensities of SERS spectra were observed primarily for hydrophobic bacterial species of Pseudomonas aeruginosa, Klebsiella pneumoniae, and Escherichia coli.
Using this technique, libraries of bacterial SERS spectra can be prepared in advance to generate reference criteria for identification of several bacterial species and strains from blood and tissue samples. The SERS biosensor can also aid caregivers in administering appropriate antibiotic treatments. After successful identification of an infecting microbe, drug resistance can be assessed by observing shifts in SERS peak intensity after incubation on antibiotic coated nanoparticles.
The Navy’s data demonstrates that SERS could not only generate unique “molecular fingerprints” for these organisms in 15-30 seconds, but could also appropriately group organisms based on commonalities. These data were confirmed by quanitiative real-time PCR samplification utilizing species primers.
Improvements to the methods of separation for sample preparation to increase bacterial extraction yields and reduce component requirements are currently underway.
This project demonstrates the valuable potential for the use of a SERS-based platform for rapid detection of microorganisms of military significance. This study sets the foundation for the utilization of a SERS platform for rapid detection of microorganism of military relevance, ultimately leading to the potential development of a field deployable point-of-care handheld detection system.
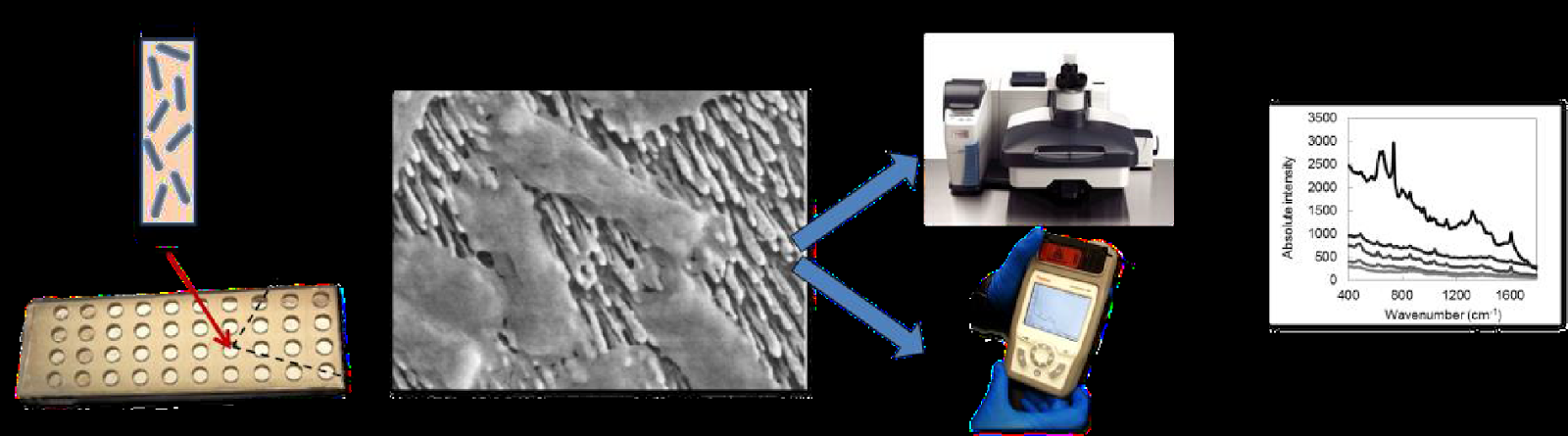
(A) Processed bacterial sample from a biological matrix inoculated into a SERS substrate well; (B) E. coli come into contact with a nanorod surface; (C) A hand-held spectrometer or desktop Raman microscope measures the SERS spectra of bacteria; (D) Unique SERS spectra of bacteria acquired utilizing SERS systems generate databases for reference purposes when diagnosing infections.
Selected Publications
Kotanen, C.N., Martinez, L., Alvarez, R., Simecek, J.W. (2016). Surface enhanced Raman scattering spectroscopy for detection and identification of microbial pathogens isolated form human serum. ScienceDirect. Vol. 8: 20-26. doi:10.1016/j.sbsr.2016.03.002
Kotanen, C., Martinez, L., Alvarez. (2015). Surface enhanced Raman spectroscopy for the rapid detection and identification of microbial pathogens in human serum. (DTIC Publication No. ADA620292). Retrieved from: www.dtic.mil/dtic/tr/fulltext/u2/a620292.pdf - 255k - 2014-12-11
Alvarez, R., Burdette, A. J., Wu, X., Kotanen, C., Zhao, Y., Tripp, R. A. (2014). Rapid identification of bacterial pathogens of military interest using surface-enhanced Raman spectroscopy. (DTIC Publication No. ADA605244). Retrieved from: www.dtic.mil/dtic/tr/fulltext/u2/a605244.pdf - 216k - 2014-06-11

